2024년 노벨 생리의학상은 Victor Ambros와 Gary Ruvkun에게 수여되었습니다. 이들은 microRNA의 발견과 그 역할에 대한 연구로 수상했으며, 특히 유전자 발현 조절에서 중요한 역할을 한다는 것을 밝혔습니다. 그들은 C. elegans라는 작은 벌레를 연구하여 microRNA가 단백질을 직접 생산하지 않고도 특정 mRNA와 결합하여 단백질 합성을 억제한다는 메커니즘을 발견했습니다. 이 연구는 유전자 조절의 새로운 원칙을 제시했고, 이후 인간을 포함한 다양한 생물체에서도 이 메커니즘이 보편적으로 적용된다는 사실이 밝혀졌습니다(NobelPrize.org).
DNA와 RNA의 구성 및 역할
- DNA (Deoxyribonucleic Acid):
- 구성성분: 이중 나선 구조, 뉴클레오타이드(당(디옥시리보스), 인산, 염기(A, T, C, G)).
- 기능: 유전 정보를 저장, 세포 내 단백질 합성 정보 제공.
- RNA (Ribonucleic Acid):
- 구성성분: 단일 나선 구조, 뉴클레오타이드(당(리보스), 인산, 염기(A, U, C, G)).
- 기능: 유전자 정보 전달 및 단백질 합성에 관여.
- 종류:
- mRNA (Messenger RNA): DNA로부터 단백질 합성 정보를 전달.
- tRNA (Transfer RNA): 아미노산을 운반하여 리보솜에서 단백질 합성.
- rRNA (Ribosomal RNA): 리보솜을 구성하고 단백질 합성 과정에 관여.
- microRNA (miRNA):
- 구성성분: 작은 RNA, 단일 가닥.
- 기능: mRNA에 결합하여 단백질 합성 억제, 유전자 발현 조절.
단위체부터 큰 단위까지:
- 뉴클레오타이드: DNA와 RNA의 기본 구성 단위.
- 염기: 아데닌(A), 구아닌(G), 사이토신(C), 티민(T, DNA), 우라실(U, RNA).
- DNA: 뉴클레오타이드들이 이중 나선 구조를 형성하며 유전 정보 저장.
- RNA: 단일 나선, 유전자 정보 전달 및 단백질 합성 관여.
https://www.nobelprize.org/prizes/medicine/2024/press-release/
The Nobel Prize in Physiology or Medicine 2024
The Nobel Prize in Physiology or Medicine 2024 was awarded jointly to Victor Ambros and Gary Ruvkun "for the discovery of microRNA and its role in post-transcriptional gene regulation"
www.nobelprize.org
Press release
2024-10-07
The Nobel Assembly at Karolinska Institutet
has today decided to award
the 2024 Nobel Prize in Physiology or Medicine
jointly to
Victor Ambros and Gary Ruvkun
for the discovery of microRNA and its role in post-transcriptional gene regulation
This year’s Nobel Prize honors two scientists for their discovery of a fundamental principle governing how gene activity is regulated.
The information stored within our chromosomes can be likened to an instruction manual for all cells in our body. Every cell contains the same chromosomes, so every cell contains exactly the same set of genes and exactly the same set of instructions. Yet, different cell types, such as muscle and nerve cells, have very distinct characteristics. How do these differences arise? The answer lies in gene regulation, which allows each cell to select only the relevant instructions. This ensures that only the correct set of genes is active in each cell type.
Victor Ambros and Gary Ruvkun were interested in how different cell types develop. They discovered microRNA, a new class of tiny RNA molecules that play a crucial role in gene regulation. Their groundbreaking discovery revealed a completely new principle of gene regulation that turned out to be essential for multicellular organisms, including humans. It is now known that the human genome codes for over one thousand microRNAs. Their surprising discovery revealed an entirely new dimension to gene regulation. MicroRNAs are proving to be fundamentally important for how organisms develop and function.
Essential regulation
This year’s Nobel Prize focuses on the discovery of a vital regulatory mechanism used in cells to control gene activity. Genetic information flows from DNA to messenger RNA (mRNA), via a process called transcription, and then on to the cellular machinery for protein production. There, mRNAs are translated so that proteins are made according to the genetic instructions stored in DNA. Since the mid-20th century, several of the most fundamental scientific discoveries have explained how these processes work.
Our organs and tissues consist of many different cell types, all with identical genetic information stored in their DNA. However, these different cells express unique sets of proteins. How is this possible? The answer lies in the precise regulation of gene activity so that only the correct set of genes is active in each specific cell type. This enables, for example, muscle cells, intestinal cells, and different types of nerve cells to perform their specialized functions. In addition, gene activity must be continually fine-tuned to adapt cellular functions to changing conditions in our bodies and environment. If gene regulation goes awry, it can lead to serious diseases such as cancer, diabetes, or autoimmunity. Therefore, understanding the regulation of gene activity has been an important goal for many decades.

The flow of genetic information from DNA to mRNA to proteins. The identical genetic information is stored in DNA of all cells in our bodies. This requires precise regulation of gene activity so that only the correct set of genes is active in each specific cell type. © The Nobel Committee for Physiology or Medicine. Ill. Mattias Karlén
In the 1960s, it was shown that specialized proteins, known as transcription factors, can bind to specific regions in DNA and control the flow of genetic information by determining which mRNAs are produced. Since then, thousands of transcription factors have been identified, and for a long time it was believed that the main principles of gene regulation had been solved. However, in 1993, this year’s Nobel laureates published unexpected findings describing a new level of gene regulation, which turned out to be highly significant and conserved throughout evolution.
Research on a small worm leads to a big breakthrough
In the late 1980s, Victor Ambros and Gary Ruvkun were postdoctoral fellows in the laboratory of Robert Horvitz, who was awarded the Nobel Prize in 2002, alongside Sydney Brenner and John Sulston. In Horvitz’s laboratory, they studied a relatively unassuming 1 mm long roundworm, C. elegans. Despite its small size, C. elegans possesses many specialized cell types such as nerve and muscle cells also found in larger, more complex animals, making it a useful model for investigating how tissues develop and mature in multicellular organisms. Ambros and Ruvkun were interested in genes that control the timing of activation of different genetic programs, ensuring that various cell types develop at the right time. They studied two mutant strains of worms, lin-4 and lin-14, that displayed defects in the timing of activation of genetic programs during development. The laureates wanted to identify the mutated genes and understand their function. Ambros had previously shown that the lin-4 gene appeared to be a negative regulator of the lin-14 gene. However, how the lin-14 activity was blocked was unknown. Ambros and Ruvkun were intrigued by these mutants and their potential relationship and set out to resolve these mysteries.

(A) C. elegans is a useful model organism for understanding how different cell types develop. (B) Ambros and Ruvkun studied the lin-4 and lin-14 mutants. Ambros had shown that lin-4 appeared to be a negative regulator of lin-14. (C) Ambros discovered that the lin-4 gene encoded a tiny RNA, microRNA, that did not code for a protein. Ruvkun cloned the lin-14 gene, and the two scientists realized that the lin-4 microRNA sequence matched a complementary sequence in the lin-14 mRNA. © The Nobel Committee for Physiology or Medicine. Ill. Mattias Karlén
After his postdoctoral research, Victor Ambros analyzed the lin-4 mutant in his newly established laboratory at Harvard University. Methodical mapping allowed the cloning of the gene and led to an unexpected finding. The lin-4 gene produced an unusually short RNA molecule that lacked a code for protein production. These surprising results suggested that this small RNA from lin-4 was responsible for inhibiting lin-14. How might this work?
Concurrently, Gary Ruvkun investigated the regulation of the lin-14 gene in his newly established laboratory at Massachusetts General Hospital and Harvard Medical School. Unlike how gene regulation was then known to function, Ruvkun showed that it is not the production of mRNA from lin-14 that is inhibited by lin-4. The regulation appeared to occur at a later stage in the process of gene expression, through the shutdown of protein production. Experiments also revealed a segment in lin-14 mRNA that was necessary for its inhibition by lin-4. The two laureates compared their findings, which resulted in a breakthrough discovery. The short lin-4 sequence matched complementary sequences in the critical segment of the lin-14 mRNA. Ambros and Ruvkun performed further experiments showing that the lin-4 microRNA turns off lin-14 by binding to the complementary sequences in its mRNA, blocking the production of lin-14 protein. A new principle of gene regulation, mediated by a previously unknown type of RNA, microRNA, had been discovered! The results were published in 1993 in two articles in the journal Cell.
The published results were initially met with almost deafening silence from the scientific community. Although the results were interesting, the unusual mechanism of gene regulation was considered a peculiarity of C. elegans, likely irrelevant to humans and other more complex animals. That perception changed in 2000 when Ruvkun’s research group published their discovery of another microRNA, encoded by the let-7 gene. Unlike lin-4, the let-7 gene was highly conserved and present throughout the animal kingdom. The article sparked great interest, and over the following years, hundreds of different microRNAs were identified. Today, we know that there are more than a thousand genes for different microRNAs in humans, and that gene regulation by microRNA is universal among multicellular organisms.
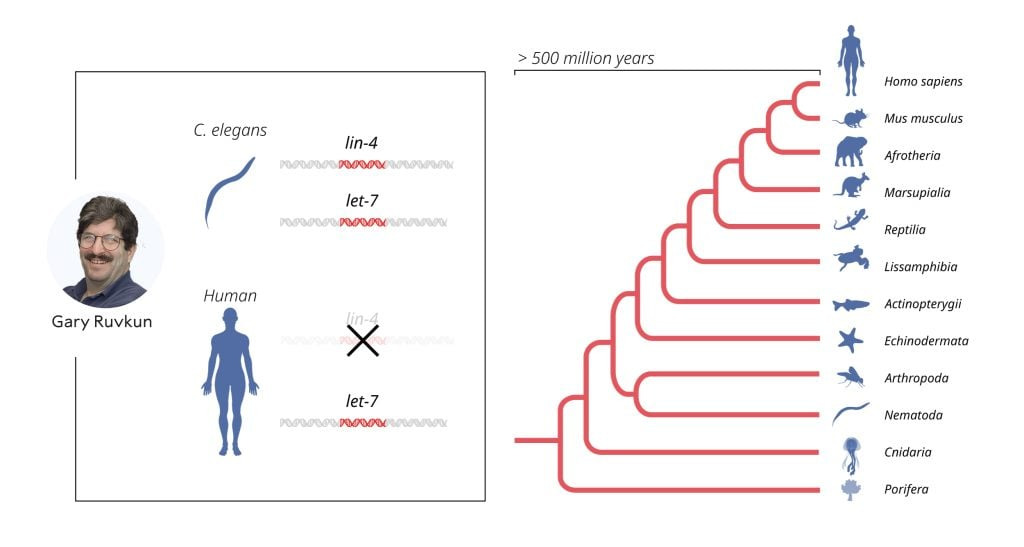
Ruvkun cloned let-7, a second gene encoding a microRNA. The gene is conserved in evolution, and it is now known that microRNA regulation is universal among multicellular organisms. © The Nobel Committee for Physiology or Medicine. Ill. Mattias Karlén
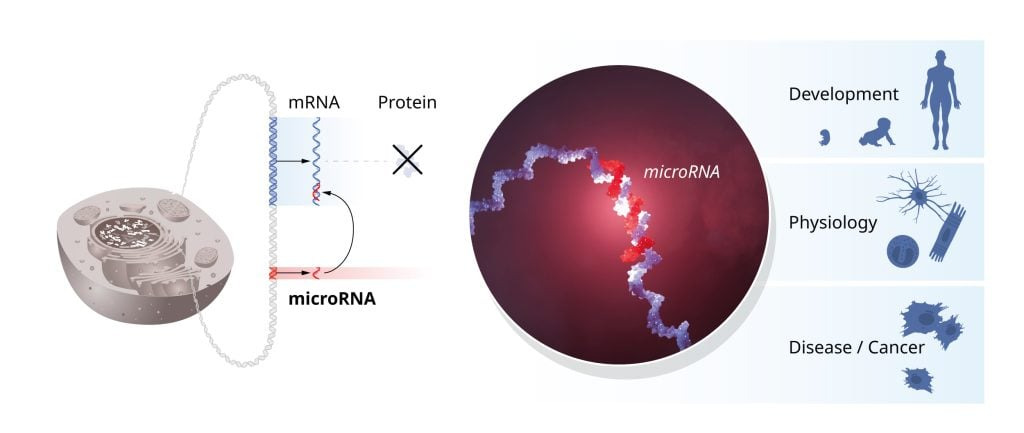
In addition to the mapping of new microRNAs, experiments by several research groups elucidated the mechanisms of how microRNAs are produced and delivered to complementary target sequences in regulated mRNAs. The binding of microRNA leads to inhibition of protein synthesis or to mRNA degradation. Intriguingly, a single microRNA can regulate the expression of many different genes, and conversely, a single gene can be regulated by multiple microRNAs, thereby coordinating and fine-tuning entire networks of genes.
Cellular machinery for producing functional microRNAs is also employed to produce other small RNA molecules in both plants and animals, for example as a means of protecting plants against virus infections. Andrew Z. Fire and Craig C. Mello, awarded the Nobel Prize in 2006, described RNA interference, where specific mRNA-molecules are inactivated by adding double-stranded RNA to cells.
Tiny RNAs with profound physiological importance
Gene regulation by microRNA, first revealed by Ambros and Ruvkun, has been at work for hundreds of millions of years. This mechanism has enabled the evolution of increasingly complex organisms. We know from genetic research that cells and tissues do not develop normally without microRNAs. Abnormal regulation by microRNA can contribute to cancer, and mutations in genes coding for microRNAs have been found in humans, causing conditions such as congenital hearing loss, eye and skeletal disorders. Mutations in one of the proteins required for microRNA production result in the DICER1 syndrome, a rare but severe syndrome linked to cancer in various organs and tissues.
Ambros and Ruvkun’s seminal discovery in the small worm C. elegans was unexpected, and revealed a new dimension to gene regulation, essential for all complex life forms.
The seminal discovery of microRNAs was unexpected and revealed a new dimension of gene regulation. © The Nobel Committee for Physiology or Medicine. Ill. Mattias Karlén
Key publications
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843-854. doi:10.1016/0092-8674(93)90529-y
Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855-862. doi:10.1016/0092-8674(93)90530-4
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kurodak MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinvasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86-89. doi:10.1038/35040556
'일상 > 후기' 카테고리의 다른 글
| 한강 작가 수상 소감 (7) | 2024.10.20 |
|---|---|
| 악이 도사리고 있을 때 (0) | 2024.10.20 |
| 10 년차 직장인의 직장생활 꿀팁 30개 (5) | 2024.10.03 |
| 에드워드 리 (Chef. Edward Lee) (1) | 2024.10.01 |
| 영국 내셔널갤러리 명화전 ‘거장의 시선, 사람을 향하다’ (0) | 2023.10.06 |